Non-local priors
Ville Karhunen
07.11.2024
nonlocalpriors.RmdNon-local densities are a family of probability distributions with zero density at the null parameter value. One of the members of this probability distribution family is a product inverse-moment (piMOM) prior for a parameter vector , with density
with parameters . The parameter controls the spread of the distribution away from the null value, and controls the tail behaviour of the distribution (Johnson, Rossell 2012). Here, we only consider the case of , which leads to Cauchy-like tails in the distribution.
The choice of can be set such that . For genetic analyses, Sanyal et al. (2019) suggested and . We can obtain the corresponding quantile from the relation of the inverse-moment prior with the (inverse) Gamma distribution (Johnson, Rossell 2010):
betaq <- 0.05
q <- 0.01
qgamma(1 - q, shape = 0.5, scale = 1)*betaq^2
#> [1] 0.008293621The marginal density with is reminiscent of the Gaussian distribution with , suggested by Wakefield (2017) and commonly used as the prior for genetic effects (e.g. Giambartolomei et al. 2014):
curve(dnorm(x, 0, 0.15), -0.8, 0.8,
col = "red", lty = 2,
xlab = bquote(beta), ylab = "Density")
curve(dimom(x, tau = 0.0083, r = 1), lty = 1, add = T)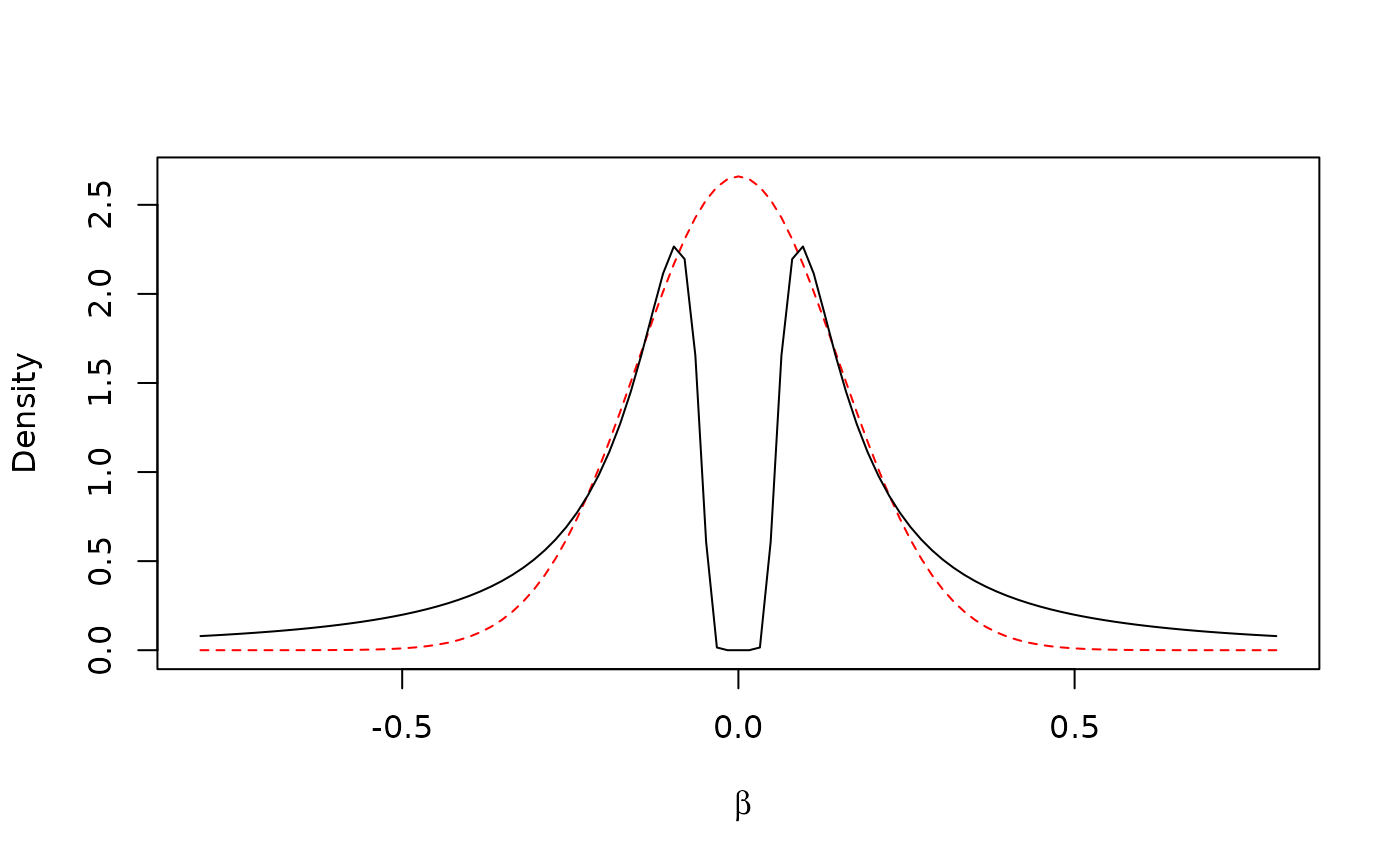
The key differences between the two distributions are that (i) the non-local prior density approaches 0 when , and (ii) the tails of the non-local density are thicker. Due to (i), non-local priors apply a strong penalty when any of its components are close to zero, therefore making them suitable for variable selection. As for (ii), the Cauchy-like tails protect from over-shrinkage of true large effects.
Note that the above example with is for illustrative purposes only – it is recommended to let be estimated based on the data (please see Karhunen et al. (2023) for details).
References
Giambartolomei et al. (2014). Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet.
Johnson, Rossell (2010). On the use of non-local prior densities in Bayesian hypothesis tests. J. R. Statist. Soc. B.
Johnson, Rossell (2012). Bayesian Model Selection in High-Dimensional Settings. J Am Stat Assoc.
Karhunen et al. (2023). Genetic fine-mapping from summary data using a nonlocal prior improves the detection of multiple causal variants. Bioinformatics.
Sanyal et al. (2019). GWASinlps: non-local prior based iterative SNP selection tool for genome-wide association studies. Bioinformatics.
Wakefield (2009). Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol.
Session information
sessionInfo()
#> R version 4.4.2 (2024-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] finimom_0.2.0
#>
#> loaded via a namespace (and not attached):
#> [1] digest_0.6.37 desc_1.4.3 R6_2.5.1 fastmap_1.2.0
#> [5] xfun_0.49 cachem_1.1.0 knitr_1.48 htmltools_0.5.8.1
#> [9] rmarkdown_2.29 lifecycle_1.0.4 cli_3.6.3 sass_0.4.9
#> [13] pkgdown_2.1.1 textshaping_0.4.0 jquerylib_0.1.4 systemfonts_1.1.0
#> [17] compiler_4.4.2 highr_0.11 tools_4.4.2 ragg_1.3.3
#> [21] evaluate_1.0.1 bslib_0.8.0 Rcpp_1.0.13-1 yaml_2.3.10
#> [25] jsonlite_1.8.9 rlang_1.1.4 fs_1.6.5