Beta-binomial prior for model dimension
Ville Karhunen
07.11.2024
betabinprior.RmdFinimom employs a beta-binomial prior for model dimension :
where is the number of variants and the maximum model size. The priors of this form with and with , are discussed in Castillo and van der Vaart (2012) and in Castillo et al. (2015). The parameter here controls the amount of prior density for smaller models, with larger values of giving more prior mass to smaller models.
Using a linkage disequilibrium (LD) matrix from an external dataset tend to increase the false positive rate. In our formulation, parameter provides a flexible way to adjust for this. The default values are when using in-sample LD matrix, and when using out-of-sample LD matrix.
We demonstrate the prior for model dimension using the example dataset:
library(finimom)
(p <- length(exampledata$betahat))
#> [1] 363
maxsize <- 10
a <- 1
u <- 1.5
val <- exp(sapply(seq_len(maxsize), dbb, p = p, a = a, b = p^u))
(val <- val/sum(val))
#> [1] 9.502616e-01 4.727099e-02 2.345333e-03 1.160564e-04 5.727772e-06
#> [6] 2.819359e-07 1.384076e-08 6.776590e-10 3.309028e-11 1.611478e-12
plot(val, type = "b", ylim = c(0, 1))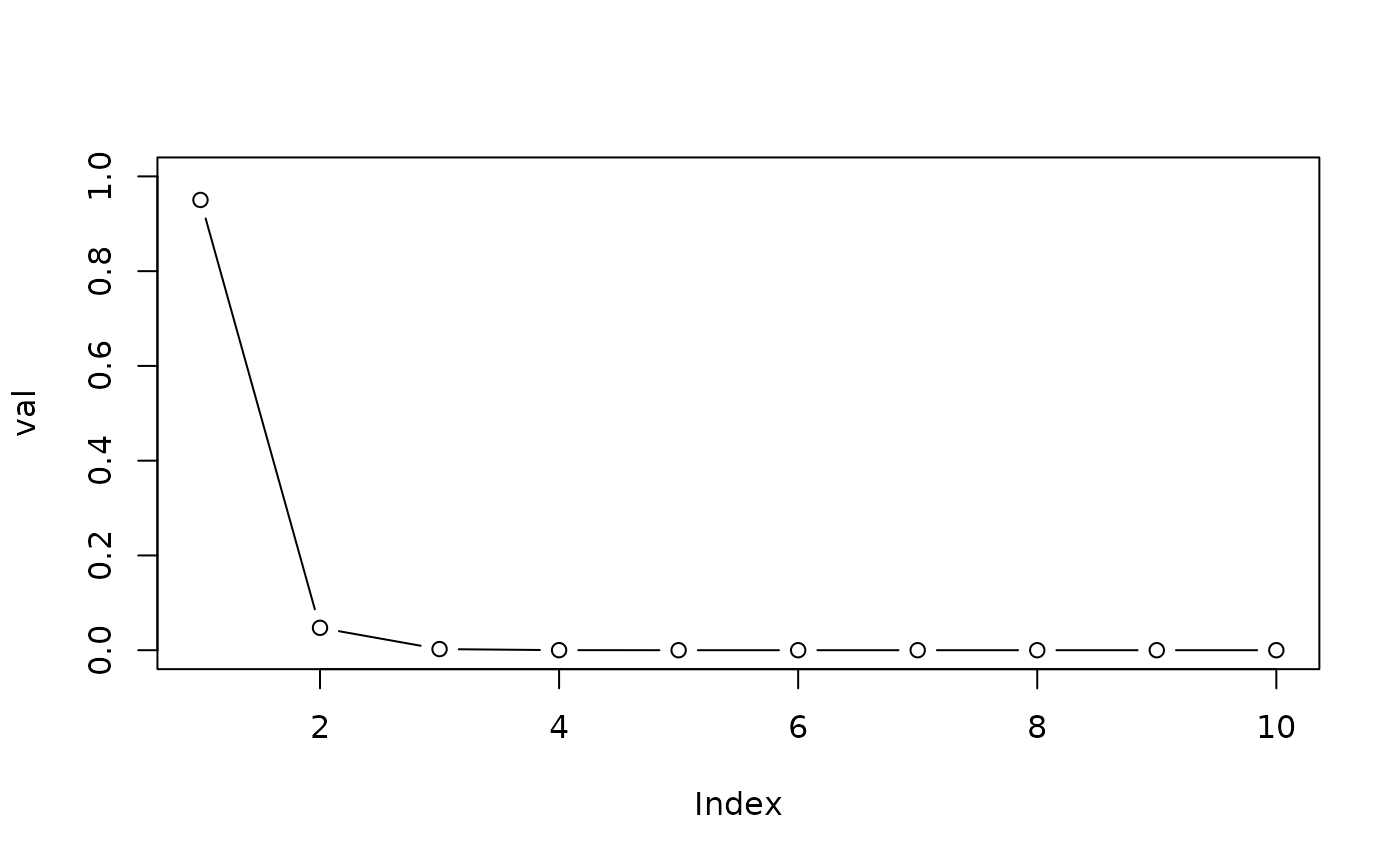
And for different values of :
us <- c(1.05, 1.5, 2, 2.25)
vals <- lapply(us, function(u){
b <- p^u
out <- exp(sapply(1:10, dbb, a = a, p = p, b = b))
out <- out/sum(out)
})
plot(vals[[1]], type = "b", ylim = c(0, 1))
invisible(lapply(2:4, function(i) lines(vals[[i]], type = "b", lty = i)))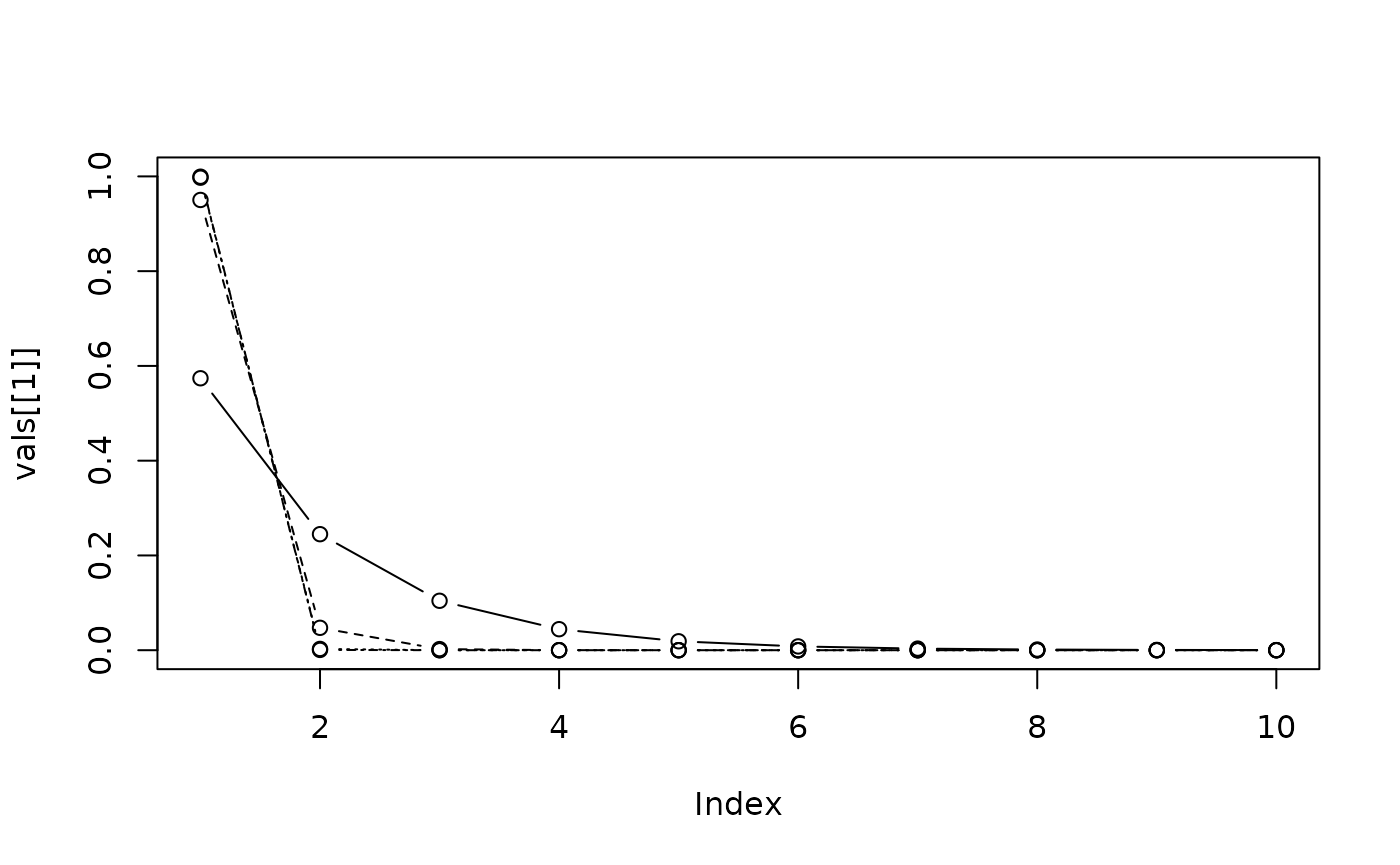
The same on a log scale:
plot(vals[[1]], type = "b", log = "y", ylim = range(unlist(vals)))
invisible(lapply(2:4, function(i) lines(vals[[i]], type = "b", lty = i)))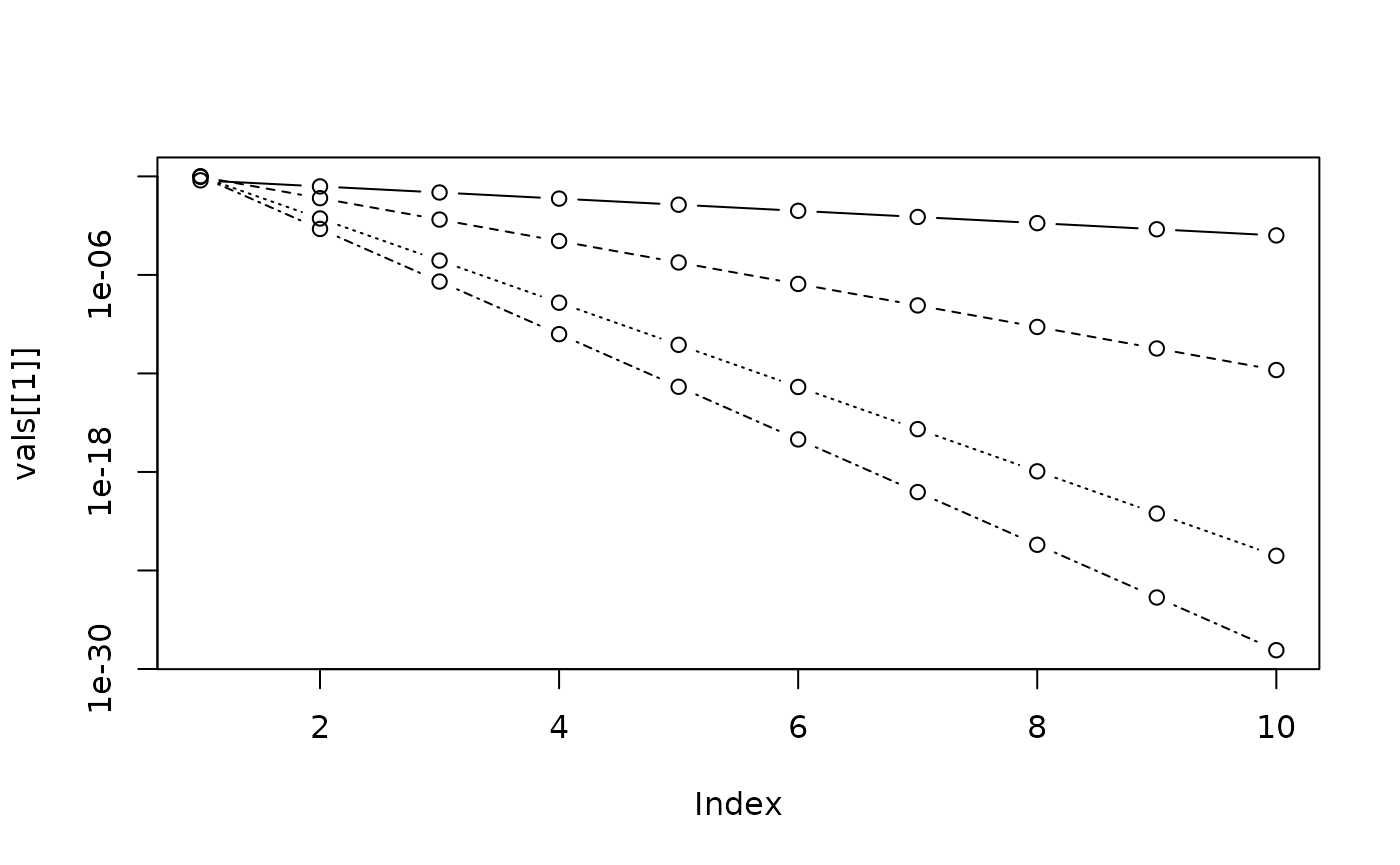
References
Castillo and van der Vaart (2012). Needles and Straw in a Haystack: Posterior concentration for possibly sparse sequences. The Annals of Statistics.
Castillo et al. (2015). Bayesian linear regression with sparse priors. The Annals of Statistics.
Session information
sessionInfo()
#> R version 4.4.2 (2024-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] finimom_0.2.0
#>
#> loaded via a namespace (and not attached):
#> [1] digest_0.6.37 desc_1.4.3 R6_2.5.1 fastmap_1.2.0
#> [5] xfun_0.49 cachem_1.1.0 knitr_1.48 htmltools_0.5.8.1
#> [9] rmarkdown_2.29 lifecycle_1.0.4 cli_3.6.3 sass_0.4.9
#> [13] pkgdown_2.1.1 textshaping_0.4.0 jquerylib_0.1.4 systemfonts_1.1.0
#> [17] compiler_4.4.2 highr_0.11 tools_4.4.2 ragg_1.3.3
#> [21] evaluate_1.0.1 bslib_0.8.0 Rcpp_1.0.13-1 yaml_2.3.10
#> [25] jsonlite_1.8.9 rlang_1.1.4 fs_1.6.5